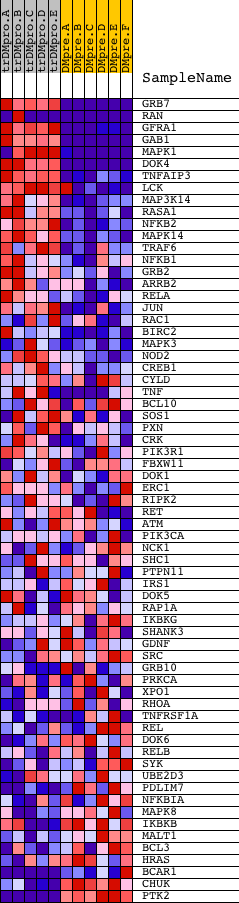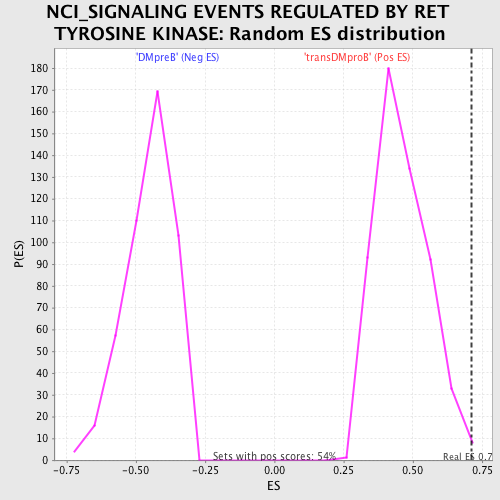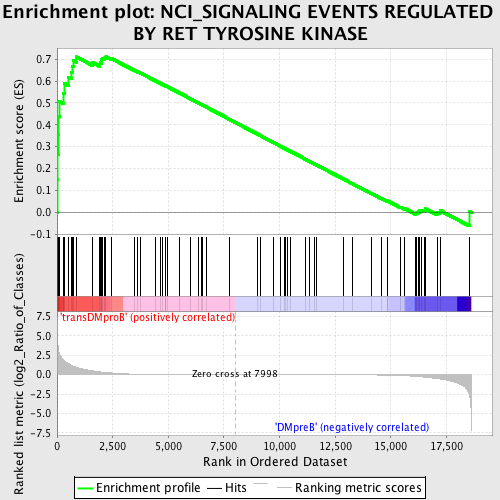
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | NCI_SIGNALING EVENTS REGULATED BY RET TYROSINE KINASE |
| Enrichment Score (ES) | 0.7104036 |
| Normalized Enrichment Score (NES) | 1.5426153 |
| Nominal p-value | 0.0036968577 |
| FDR q-value | 0.16139211 |
| FWER p-Value | 0.834 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 5 | 5.748 | 0.1507 | Yes | ||
| 2 | RAN | 5356 9691 | 22 | 4.446 | 0.2665 | Yes | ||
| 3 | GFRA1 | 9015 | 56 | 3.423 | 0.3546 | Yes | ||
| 4 | GAB1 | 18828 | 64 | 3.185 | 0.4379 | Yes | ||
| 5 | MAPK1 | 1642 11167 | 108 | 2.667 | 0.5056 | Yes | ||
| 6 | DOK4 | 18785 | 290 | 1.879 | 0.5451 | Yes | ||
| 7 | TNFAIP3 | 19810 | 331 | 1.761 | 0.5892 | Yes | ||
| 8 | LCK | 15746 | 521 | 1.410 | 0.6160 | Yes | ||
| 9 | MAP3K14 | 11998 | 645 | 1.197 | 0.6408 | Yes | ||
| 10 | RASA1 | 10174 | 698 | 1.106 | 0.6671 | Yes | ||
| 11 | NFKB2 | 23810 | 744 | 1.056 | 0.6924 | Yes | ||
| 12 | MAPK14 | 23313 | 872 | 0.931 | 0.7100 | Yes | ||
| 13 | TRAF6 | 5797 14940 | 1581 | 0.489 | 0.6847 | Yes | ||
| 14 | NFKB1 | 15160 | 1890 | 0.360 | 0.6775 | Yes | ||
| 15 | GRB2 | 20149 | 1927 | 0.347 | 0.6847 | Yes | ||
| 16 | ARRB2 | 20806 | 1981 | 0.330 | 0.6905 | Yes | ||
| 17 | RELA | 23783 | 1985 | 0.329 | 0.6990 | Yes | ||
| 18 | JUN | 15832 | 2037 | 0.313 | 0.7045 | Yes | ||
| 19 | RAC1 | 16302 | 2149 | 0.281 | 0.7059 | Yes | ||
| 20 | BIRC2 | 4397 4398 | 2196 | 0.267 | 0.7104 | Yes | ||
| 21 | MAPK3 | 6458 11170 | 2440 | 0.214 | 0.7029 | No | ||
| 22 | NOD2 | 6384 | 3468 | 0.062 | 0.6492 | No | ||
| 23 | CREB1 | 3990 8782 4558 4093 | 3625 | 0.050 | 0.6421 | No | ||
| 24 | CYLD | 18532 | 3768 | 0.043 | 0.6356 | No | ||
| 25 | TNF | 23004 | 4411 | 0.021 | 0.6015 | No | ||
| 26 | BCL10 | 15397 | 4654 | 0.016 | 0.5889 | No | ||
| 27 | SOS1 | 5476 | 4722 | 0.015 | 0.5857 | No | ||
| 28 | PXN | 5339 3573 | 4852 | 0.013 | 0.5791 | No | ||
| 29 | CRK | 4559 1249 | 4892 | 0.013 | 0.5773 | No | ||
| 30 | PIK3R1 | 3170 | 4940 | 0.013 | 0.5751 | No | ||
| 31 | FBXW11 | 20926 | 5509 | 0.008 | 0.5447 | No | ||
| 32 | DOK1 | 17104 1018 1177 | 5995 | 0.006 | 0.5187 | No | ||
| 33 | ERC1 | 1013 17021 995 1136 | 6373 | 0.004 | 0.4985 | No | ||
| 34 | RIPK2 | 2528 15935 | 6498 | 0.004 | 0.4919 | No | ||
| 35 | RET | 17028 | 6549 | 0.004 | 0.4893 | No | ||
| 36 | ATM | 2976 19115 | 6723 | 0.003 | 0.4801 | No | ||
| 37 | PIK3CA | 9562 | 7760 | 0.001 | 0.4242 | No | ||
| 38 | NCK1 | 9447 5152 | 8993 | -0.002 | 0.3579 | No | ||
| 39 | SHC1 | 9813 9812 5430 | 9137 | -0.003 | 0.3503 | No | ||
| 40 | PTPN11 | 5326 16391 9660 | 9713 | -0.004 | 0.3194 | No | ||
| 41 | IRS1 | 4925 | 10028 | -0.005 | 0.3026 | No | ||
| 42 | DOK5 | 2945 14722 | 10205 | -0.006 | 0.2932 | No | ||
| 43 | RAP1A | 8467 | 10259 | -0.006 | 0.2905 | No | ||
| 44 | IKBKG | 2570 2562 4908 | 10346 | -0.006 | 0.2861 | No | ||
| 45 | SHANK3 | 22385 404 | 10507 | -0.006 | 0.2776 | No | ||
| 46 | GDNF | 22514 2275 | 11186 | -0.009 | 0.2413 | No | ||
| 47 | SRC | 5507 | 11341 | -0.009 | 0.2332 | No | ||
| 48 | GRB10 | 4799 | 11584 | -0.010 | 0.2204 | No | ||
| 49 | PRKCA | 20174 | 11674 | -0.011 | 0.2159 | No | ||
| 50 | XPO1 | 4172 | 12872 | -0.020 | 0.1519 | No | ||
| 51 | RHOA | 8624 4409 4410 | 13271 | -0.024 | 0.1311 | No | ||
| 52 | TNFRSF1A | 1181 10206 | 14144 | -0.040 | 0.0851 | No | ||
| 53 | REL | 9716 | 14584 | -0.054 | 0.0629 | No | ||
| 54 | DOK6 | 509 | 14833 | -0.066 | 0.0512 | No | ||
| 55 | RELB | 17942 | 14836 | -0.066 | 0.0529 | No | ||
| 56 | SYK | 21636 | 15422 | -0.112 | 0.0243 | No | ||
| 57 | UBE2D3 | 7253 | 15623 | -0.136 | 0.0171 | No | ||
| 58 | PDLIM7 | 7434 | 16120 | -0.220 | -0.0039 | No | ||
| 59 | NFKBIA | 21065 | 16172 | -0.230 | -0.0006 | No | ||
| 60 | MAPK8 | 6459 | 16226 | -0.245 | 0.0029 | No | ||
| 61 | IKBKB | 4907 | 16287 | -0.259 | 0.0065 | No | ||
| 62 | MALT1 | 6274 | 16378 | -0.279 | 0.0090 | No | ||
| 63 | BCL3 | 8654 | 16506 | -0.311 | 0.0103 | No | ||
| 64 | HRAS | 4868 | 16569 | -0.329 | 0.0156 | No | ||
| 65 | BCAR1 | 18741 | 17079 | -0.500 | 0.0013 | No | ||
| 66 | CHUK | 23665 | 17214 | -0.556 | 0.0087 | No | ||
| 67 | PTK2 | 22271 | 18532 | -2.547 | 0.0045 | No |